Research Projects
Overview
Mass spectrometry (MS) has proven to be a powerful analytical tool in many scientific fields for nearly a century. Advances made in soft ionization techniques such as electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI) have expanded the use of MS to the biological sciences, allowing biomolecules to be ionized and sampled in the gas phase without fragmentation. These advances have been echoed in an explosion of publications over the past 20 years that have used MS as a tool to illuminate information pertaining to a biological question. The majority of MS-based bioanalytical studies to date have focused on cataloguing the species present in a sample at a static moment in time. Although this is important, measuring the dynamics of a system in response to a stimulus not only aids identification efforts, but offers mechanistic insight into the cellular processes involved. Gaining a better understanding of these dynamics, often expressed through a change in concentration or in the degree of modification, will enhance the accuracy of cellular models, assign functional roles to the identified species as well as highlight new diagnostic and therapeutic strategies. The research program of the Smith group will focus on developing novel methods to efficiently measure biomolecular dynamics in living systems with the downstream goals of elucidating disease biomarkers and further characterizing biosynthetic and biochemical pathways.
Instrumentation
The Smith lab uses mass spectrometry as a tool to probe the dynamics of various biomolecules. At present, we mainly conduct research in the area of protein analysis (“proteomics”) and lipid analysis (“lipidomics”). The laboratory is equipped with high-resolution hybrid quadrupole time of flight, triple quadrupole and linear ion trap mass spectrometers. All of these instruments achieve ionization via electrospray ionization and are coupled to high performance liquid chromatographs. The lab additionally houses two gas chromatography-MS systems, one equipped with a headspace sampler.
TrEnDi
Trimethylation Enhancement using Diazomethane (TrEnDi) is a published technique developed by Professors Jeffrey Smith and Jeffrey Manthorpe at Carleton University. The method increases the sensitivity of mass spectrometric detection by assigning a fixed, permanent positive charge to amino groups. It allows for increased and predictable sequence coverage for peptides in proteomic analyses, and increased limits of detection for several important lipid classes in lipidomic analyses.
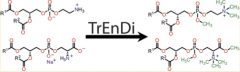
Quantitative Proteomics
The Smith lab uses microfluidic devices to manipulate biological samples in a highly efficient manner to elucidate protein dynamics and post-translational modification patterns using MS in conjunction with TrEnDi. Through studying the dynamics of protein abundances or the modes in which they are modified in cells that are stimulated in some manner (or that are either diseased or healthy), our research will help define how cells communicate with each other, their environment and themselves.
Quantitative Lipidomics
Although lipids have been studied for decades and have been largely regarded as energy storage or structural molecules, recent progress in lipid research has revealed many novel and important roles for lipids in cellular signaling. Many classes of lipids are easily analyzed by mass spectrometry; however, some are more difficult to observe. The Smith laboratory will focus on developing novel methods to identify lipid species in the context of complicated biological samples. To date, the development of TrEnDi has achieved this goal by increasing the sensitivity of some lipid classes over an order of magnitude. Ultimately, we will apply the novel methods that are developed to investigate lipid dynamics in biological systems to aid our understanding of the roles they play in cellular life.