Check out the Maria’s Google Scholar page for the latest information about the DeRosa lab publications.
Patent Applications
APTAMERS AS A THERAPEUTIC TOOL TO PREVENT PROTEIN AGGREGATION IN NEURODEGENERATIVE DISEASE DEROSA, Maria Cynthia; HOLAHAN, Matthew Richard; MCCONNELL, Erin Marie; VENTURA, Katelyn Victoria; CALLAHAN, Joshua Parker; HUNT, Vernon Harold Daniel US Patent Application 62/575,813 https://patents.google.com/patent/WO2019079887A1/en
ROOT EXUDATE-ACTIVATED SYSTEM FOR AGROCHEMICAL DELIVERY SCHNEIDER, Juan; MONREAL, Carlos; DEROSA, Maria; CHOI, Phillip; MASTRONARDI, Emily; TSAE, Phepafatso; MATUS, Francisco. PCT/CA2019/051306 https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2020051717
Publication list 2018-2024 (Bold indicates LADDER HQP)
113) E.M. McConnell, D. Chan, K. Ventura, J.P. Callahan, K. Harris, V.H. Hunt, S. Boisjoli, D. Knight, E.T. Monk, M.R. Holahan, M.C. DeRosa, “Selection of DNA aptamers that prevent the fibrillization of α-synuclein protein in cellular and mouse models” Molec. Ther. Nucleic Acids. 2024. DOI: https://doi.org/10.1016/j.omtn.2024.102251

112) B. Chovelon, V. Ranganathan, S. Srinivasan, E.M. McConnell, P. Faure, E. Fiore, C. Ravelet, E. Peyrin, M.C. DeRosa, “Noncompetitive determination of small analytes by Sandwich-type lateral flow assay based on aptamer kissing complex” Anal. Chem. 2024. 96, 18, 6875-6880. DOI: https://doi.org/10.1021/acs.analchem.3c05472
111) S.A. Tittlemier, B. Cramer, M.C. DeRosa, Z. Dzuman, R. Malone, C. Maragos, M. Suman, M.W. Sumarah, “Developments in analytical techniques for mycotoxin determination: an update for 2022-2023” World Mycotoxin Journal. 2024. 17(1), 3-26, https://doi.org/10.1163/18750796-bja10002
110) B. Billet, B. Chovelon, E.M. McConnell, D. Andre, L. Puillet-Anselme, E. Fiore, P. Faure, C. Ravelet, M.C. DeRosa, E. Peyrin, “Iodinated organic molecule as tag for inductively-coupled plasma-mass spectrometry, aptamer assays” Talanta. 2024. DOI: https://doi.org/10.1016/j.talanta.2023.125107
109) F. Ebanks, H. Nasrallah, T.M. Garant, E.M. McConnell, M.C. DeRosa, “Colorimetric detection of aflatoxins B1 and M1 using aptamers and gold and silver nanoparticles” Advanced Agrochem. 2023. 2, 221. https://doi.org/10.1016/j.aac.2023.07.003
108) R.S. Massey, E.M. McConnell, D. Chan, M.R. Holahan, M.C. DeRosa, R. Prakash, “Non-invasive Monitoring of α-Synuclein in Saliva for Parkinson’s Disease Using Organic Electrolyte-Gated FET Aptasensor” ACS Sensors. 2023. 8, 3116. https://doi.org/10.1021/acssensors.3c00757
107) M.C. DeRosa, A. Lin, P. Mallikaratchy, E.M. McConnell, M. McKeague, Rutika Patel, Sarah Shigdar, “In vitro selection of aptamers and their applications” Nat. Rev. Methods Primers. 2023, 3, 54. https://doi.org/10.1038/s43586-023-00238-7
106) S. Srinivasan, V. Ranganathan, E.M. McConnell, B.M.M. Murari, M.C. DeRosa, “Aptamer-based colorimetric and lateral flow assay approaches for the detection of toxic metal ions, thallium (i) and lead (ii)” RSC Adv. 2023. 13, 20040. https://doi.org/10.1039/D3RA01658G
105) M. Pundir, S. Papagerakis, M. C. De Rosa, N. Chronis, K. Kurabayashi, S. Abdulmawjood, M. E. P. Prince, L. Lobanova, X. Chen, “Emerging biotechnologies for evaluating disruption of stress, sleep, and circadian rhythm mechanism using aptamer-based detection of salivary biomarkers” Biotech. Advances. 2022, 59, 107961. https://doi.org/10.1016/j.biotechadv.2022.107961
104) V. Ranganathan, S. Boisjoli, M. C. DeRosa* “Adsorption–Desorption Nano-Aptasensors: Fluorescent screening assays for Ochratoxin A” RSC Advances. 2022 12, 13727-13739. https://doi.org/10.1039/D2RA00026A
103) S.A. Tittlemier, B. Cramer, C. Dall’Asta, M.C. DeRosa, V.M.T. Lattanzio, R. Malone, C. Maragos, M. Stranska, M.W. Sumarah “Developments in mycotoxin analysis: an update for 2020-21” World Mycotoxin Journal 2022, 15, 3-25. https://doi.org/10.3920/WMJ2021.2752
102) A. Koudrina, C. Chartrand, G. O. Cron, J. O’Brien, E. C. Tsai, M. C. DeRosa* “Fibrinogen aptamer functionalized gold-coated iron-oxide nanoparticles for targeted imaging of thrombi” Chem. Comm. 2022, 58, 2870-2873. https://doi.org/10.1039/D1CC03817F
101) M. Loyez*, M. C. DeRosa, C. Caucheteur, R. Wattiez “Overview and emerging trends in optical fiber aptasensing” Biosens. Bioelectron. 2022, 196, 113694. https://doi.org/10.1016/j.bios.2021.113694
100) Z. Abdulsada, R. Kibbee, D. Schwertfeger, J. Princz, M. DeRosa, B. Örmeci* “Fate and Removal of Silver Nanoparticles during Sludge Conditioning and their Impact on Soil Health after Simulated Land Application” Water Res. 2021, 206, 117757. https://doi.org/10.1016/j.watres.2021.117757
99) K. G. Earnest, E. M. McConnell, E. M. Hassan, M. Wunderlich, B. Hosseinpour, B. S. Bono, M. J. Chee, J. C. Mulloy, W. G. Willmore, M. C. DeRosa*, E. J. Merino* “Development and characterization of a DNA aptamer for MLL-AF9 expressing acute myeloid leukemia cells using whole cell-SELEX” Sci. Rep. 2021, 11, 19174. https://doi.org/10.1038/s41598-021-98676-4
98) Z. Abdulsada, R. Kibbee, J. Princz, M. C. DeRosa, B. Örmeci* “Transformation of Silver Nanoparticles (AgNPs) during Lime Treatment of Wastewater Sludge and Their Impact on Soil Bacteria” Nanomaterials 2021, 11, 2330. https://doi.org/10.3390/nano11092330
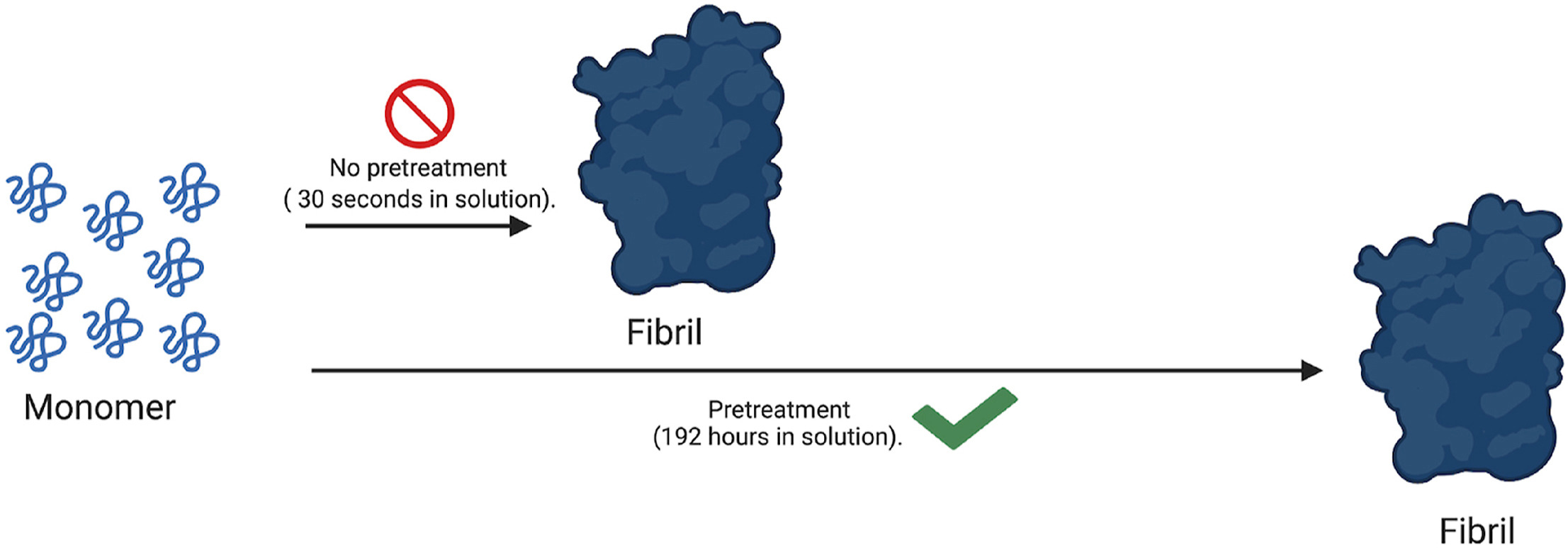
97) M. Q. Ferguson, M.C. DeRosa* “Optimized experimental pre-treatment strategy for temporary inhibition of islet amyloid polypeptide aggregation”. Biochem Biophys Rep. 2021, 26, 100964. https://doi.org/10.1016/j.bbrep.2021.100964
96) S. Natarajan*, M. C. DeRosa, M.I. Shah, J. Joseph. “Development and evaluation of a quantitative fluorescent lateral flow immunoassay for cystatin C, a renal dysfunction biomarker.” Sensors. 2021, 21, 3178. https://doi.org/10.3390/s21093178
95) J.R. Velicogna, D. Schwertfeger, A. Jesmer, C. Beer, J. Kuo, M. C. DeRosa, R. Scroggins, M. Smith, J. Princz*. “Soil invertebrate toxicity and bioaccumulation of nano copper oxide and copper sulphate in soils, with and without biosolids amendment.” Ecotox Environ Safety. 2021, 217, 112222. https://doi.org/10.1016/j.ecoenv.2021.112222
94) D. Marquez, J. Chawich, W. Hassen, K. Moumanis, J. Dubowski*, M.C. DeRosa*. “Polymer Brushes-GaAs Interface and its use as an Antibody-compatible Platform for Biosensing.” ACS Omega 2021, 6, 7286-7295. https://doi.org/10.1021/acsomega.0c04954
93) M. Loyez, M. Lobry, E. M. Hassan, M. C. DeRosa, C. Caucheteur, R. Wattiez*. “HER2 breast cancer biomarker detection using a sandwich optical fiber assay” Talanta 2021, 121452. https://doi.org/10.1016/j.talanta.2020.121452

92) E. M. Hassan, B. R. Dixon, S. A. Sattar, A. Stalker, B. Örmeci, M. C. DeRosa*. “Highly sensitive magnetic microparticle-based aptasensor for Cryptosporidium parvum oocyst detection in river water and wastewater: Effect of truncation on aptamer affinity” Talanta, 2021, 222, 121618. https://doi.org/10.1016/j.talanta.2020.121618
91) Z. K. Abdulsada, R. Kibbee, B. Örmeci, M. DeRosa, J. Princz*. “Impact of anaerobically digested silver and copper oxide particles in biosolids on soil characteristics and bacterial community” Chemosphere 2021 263, 128173. https://doi.org/10.1016/j.chemosphere.2020.128173
90) S.A. Tittlemier*, J. Brunkhorst, B. Cramer, M.C. DeRosa, V.M.T. Lattanzio, R. Malone, C. Maragos, M. Stranska, M.W. Sumarah. “Developments in mycotoxin analysis: an update for 2019-20” World Mycotoxin Journal 2021 14, 3-26. https://doi.org/10.3920/WMJ2020.2664 (Invited)
89) D. Shimwe Niyonambaza, A. Koudrina, M. C. DeRosa, M. Boukadoum, A. Miled, E. Boisselier* “Aptamer-Modified Ultrastable Gold Nanoparticles for Dopamine Detection” IEEE Sensors 2021, 21, 2517-2525. doi: 10.1109/JSEN.2020.3026159.
88) A. Koudrina, E. M. McConnell, J. A. Zurakowski, G. O. Cron, S. Chen, E. Tsai, M. C. DeRosa* “Developing aptamer-conjugated gadolinium contrast media to target fibrin clots in magnetic resonance imaging” ACS Applied Mat Inter. 2021, 13, 8, 9412-9424. https://doi.org/10.1021/acsami.0c16666
87) E. M. Hassan, B. Örmeci, M.C. DeRosa*, B. R. Dixon, S. A. Sattar*, A. Iqbal. “A review of Cryptosporidium spp. and their detection in water.” Water Sci. Tech. 2021, 83, 1-25. https://doi.org/10.2166/wst.2020.515
86) E. Mastronardi, K. Cyr, C. Monreal, M.C. DeRosa*. “Selection of DNA aptamers for root exudate L- serine using multiple selection strategies for use in agricultural biosensing”. J. Ag. Food Chem. 2021, 69, 4294-4306. https://doi.org/10.1021/acs.jafc.0c06796 Cover article

85) C. Kolm, I. Cervenka, U. J. Aschl, N. Baumann, S. Jakwerth, R. Krska, R. L.Mach, R. Sommer, M.C.DeRosa, A.K.T. Kirschner, A.H. Farnleitner, G.H. Reisch*. “DNA aptamers against bacterial cells can be efficiently selected by a SELEX process using state-of-the-art qPCR and ultra-deep sequencing” Sci. Rep. 2020, 10, 20917. https://doi.org/10.1038/s41598-020-77221-9
84) M. Lobry, M. Loyez*, K. Chah, E. M. Hassan, E. Goormaghtigh, M. C. DeRosa, R. Wattiez, C Caucheteur*. “HER2 biosensing through SPR-envelope tracking in plasmonic optical fiber gratings”. Biomed. Opt. Express. 2020, 11, 4862-4871. https://doi.org/10.1364/BOE.401200
83) M. Lobry, M. Loyez, E. M. Hassan, K. Chah, M. C. DeRosa, E. Goormaghtigh, R. Wattiez, C. Caucheteur* “Multimodal plasmonic optical fiber grating aptasensor” Optics Express, 2020, 28, 7539-7551. https://doi.org/10.1364/OE.385747.
82) M. Loyez, E. M. Hassan, M. Lobry, F. Liu, C. Caucheteur, R. Wattiez, M. C. DeRosa, W. G. Willmore, J. Albert* “Rapid detection of circulating breast cancer cells using a multi-resonant optical fiber aptasensor with plasmonic amplification” ACS Sens. 2020, 5, 2, 454-463. https://doi.org/10.1021/acssensors.9b02155.
81) E. M. Hassan, M. C. DeRosa* “Recent advances in cancer early detection and diagnosis: role of nucleic acid based aptasensors” TrAC Trends in Anal. Chem. 2020, 124, 115806. https://doi.org/10.1016/j.trac.2020.115806.
80) R. Velu, S. Srinivasan, A. Singh, M. C. DeRosa* “An aptamer-based colorimetric lateral flow assay for the detection of human epidermal growth factor receptor 2 (HER2)” Anal Biochem. 2020, 588, 113471, https://doi.org/10.1016/j.ab.2019.113471.
79) M. Loyez, E. Hassan, M. Lobry, F. Liu, C. Caucheteur, R. Wattiez, M. DeRosa, W. Willmore, J. Albert.* “Circulating cancer cell detection using an optical fiber aptasensor”. Proc. SPIE Biophotonics in Point-of-Care. 2020, 113610. https://doi.org/10.1117/12.2557775
78) N.M. Sopinka*, L. E. Coristine, M. C. DeRosa, C. M. Rochman, B. L. Owens, S. J. Cooke. “Envisioning the scientific paper of the future” FACETS 2020, 5, 1-16. https://doi.org/10.1139/facets-2019-0012
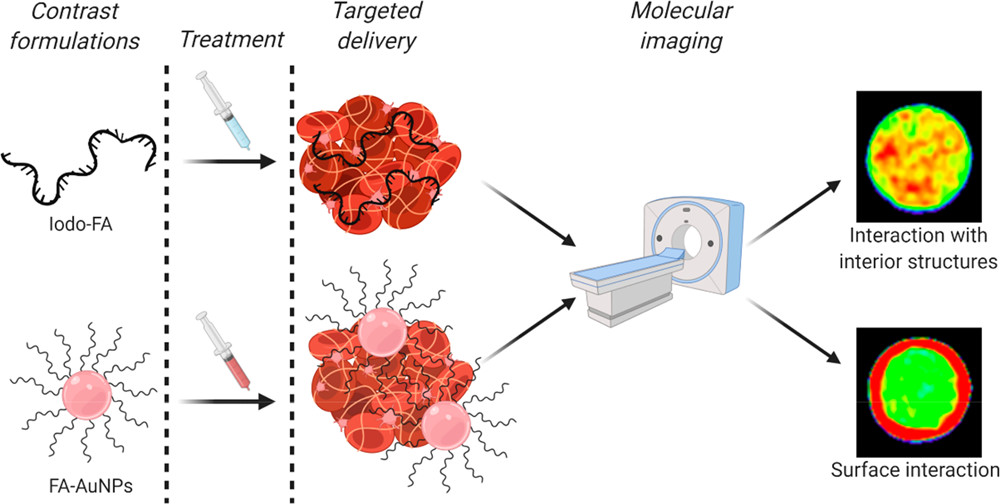
77) A. Koudrina, J. O’Brien, R. Garcia, S. Boisjoli, P. Kan, E. Tsai, M. C. DeRosa*. “Assessment of aptamer targeted contrast agents for monitoring of blood clots in computed tomography and fluoroscopy imaging.” Bioconj. Chem 2020, 31, 2737-2749. https://doi.org/10.1021/acs.bioconjchem.0c00525
76) F. Ciriaco, V. De Leo, L. Catucci, M. Pascale, A. F. Logrieco, M. C. DeRosa, A. De Girolamo*. “An in silico pipeline for rapid screening of DNA aptamers against mycotoxins: The case study of Fumonisin B1, Aflatoxin B1, and Ochratoxin A” Polymers 2020, 12, 2983. https://doi.org/10.3390/polym12122983
75) A. Koudrina, M.C. DeRosa*. “Advances in Medical Imaging: Aptamer- and Peptide-Targeted MRI and CT Contrast Agents.” ACS Omega 2020, 5, 22691-22701. https://doi.org/10.1021/acsomega.0c02650 (Invited)
74) S. Mohan, J. Princz, B. Ormeci, M. C. DeRosa* “Morphological transformation of silver nanoparticles from commercial products: Modeling from product incorporation, weathering through use scenarios, and leaching into wastewater” Nanomaterials 2019, 9 (9), 1258. https://doi.org/10.3390/nano9091258.
73) R. Massey, E. Morin, M. C. DeRosa, R. Prakash* Label-free Detection of Dopamine Using Aptamer Enhanced Organic-Electrolyte Gated FET Sensor. 2019, 2019 IEEE International Conference on Flexible and Printable Sensors and Systems, DOI: 10.1109/FLEPS.2019.8792324
72) S. Chen, A. M. Auriat, T. Li, T. R. Stumpf, R.Wylie, X. Chen, S. M. Willerth, M.C. DeRosa, M. Tarizian, X. Cao, E. C. Tsai* Advancements in Canadian Biomaterials in Neurotraumatic Diagnosis and Therapies. Processes 2019, 7 (6), 336. https://doi.org/10.3390/pr7060336
71) S. Srinivasan, V. Ranganathan, M.C. DeRosa, B.M. Murari* Comparison of turn-on and ratiometric fluorescent G-quadruplex aptasensor approaches for the detection of ATP. Anal. Bioanal. Chem. 2019, 1-12. https://doi.org/10.1007/s00216-018-1484-x Front Cover

70) E. M. Hassan, A. Mohamed, M. C. DeRosa, W. G. Willmore, Y. Hanaoka, T. Kiwa, T. Ozaki*. High-sensitivity detection of metastatic breast cancer cells via terahertz chemical microscopy using aptamers. Sens. Act. B. Chem. 2019, 287, 595-601. https://doi.org/10.1016/j.snb.2019.02.019
69) M. Smith, M. C. DeRosa*, Synthesis, Transfer and Characterization of Core-Shell Gold-Coated Magnetic Nanoparticles. MethodsX, 2019, 6, 333-354. https://doi.org/10.1016/j.mex.2019.02.006
68) E. M. McConnell, K. Ventura, Z. Dwyer, M. R. Holahan, M.C. DeRosa* In vivo use of a multi-DNA aptamer-based payload/targeting system to study dopamine dysregulation in the central nervous system. ACS Chem. Neuro. 2019, 10, 371-383. https://doi.org/10.1021/acschemneuro.8b00292
67) M. Belleperche, M. C. DeRosa* pH-control in aptamer-based diagnostics, therapeutics, and analytical applications. Pharmaceuticals 2018, 11, 80 – 93. https://doi.org/10.3390/ph11030080
66) R. Velu, M. C. DeRosa*, Lateral Flow Assays for Ochratoxin A: Comparison of Linkage inversion assembled nano-aptasensors (LIANA) and label-free approaches. Analyst, 2018, 143, 4566-4574. Doi: 10.1039/C8AN00963E Back Cover and Hot Article

65) S. Srinivasan, R. Velu, M. C. DeRosa, and B. M. Murari* Label-free aptasensor based on a fluorescent screening assay for the detection of Salmonella typhimurium. Analytical biochem. 2018, 559, 17-23. https://doi.org/10.1016/j.ab.2018.08.002
64) C. M. Monreal*, J. Zhang, S. Koziel, J. Vidmar, M. González, F. Matus, S. Baxi, S. Wu, M. C. DeRosa, P. Etcheverria “Bacterial community structure associated with the addition of nitrogen and the dynamics of soluble carbon in the rhizosphere of canola (Brassica napus) grown in a Podzol” Rhizosphere, 2018, 5, 16-25. https://doi.org/10.1016/j.rhisph.2017.11.004
63) E. Mastronardi, C. Monreal, M. C. DeRosa* “Personalized medicine for crops? Opportunities for the application of molecular recognition in agriculture” J. Agri. Food. Chem 2018, 66: 6457-6461. DOI: 10.1021/acs.jafc.7b03295

Publication list 2009-2017
62) A. Ruscito, E. M. McConnell, A. Koudrina, R. Velu, C. Mattice, V. Hunt, M. McKeague*, and M. C. DeRosa* “In Vitro Selection and Characterization of DNA Aptamers to a Small Molecule Target” Current Protocols in Chemical Biology, 2017, 9, 1–36. DOI 10.1002/cpch.28
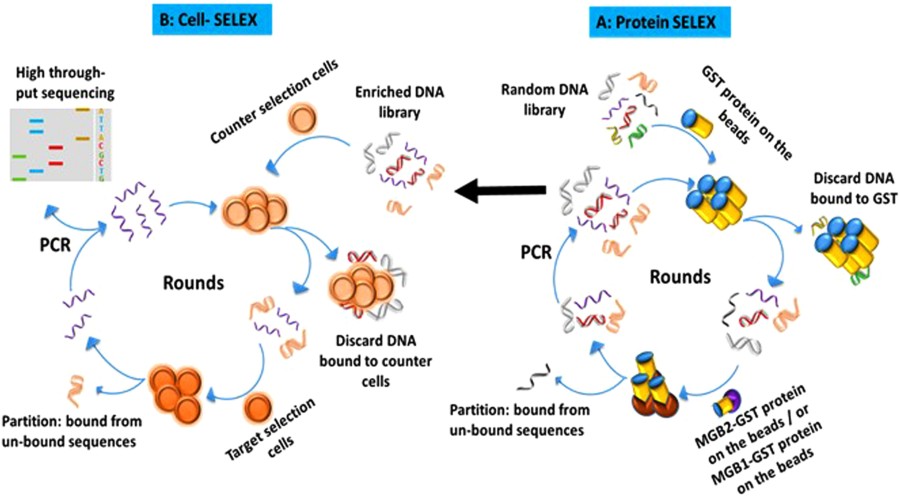
61) E. M. Hassan, W. Willmore, M. C. DeRosa* “In vitro selections of mammaglobin A and mammaglobin B aptamers for the recognition of circulating breast tumor cells” Sci. Rep 2017,doi:10.1038/s41598-017-13751-z
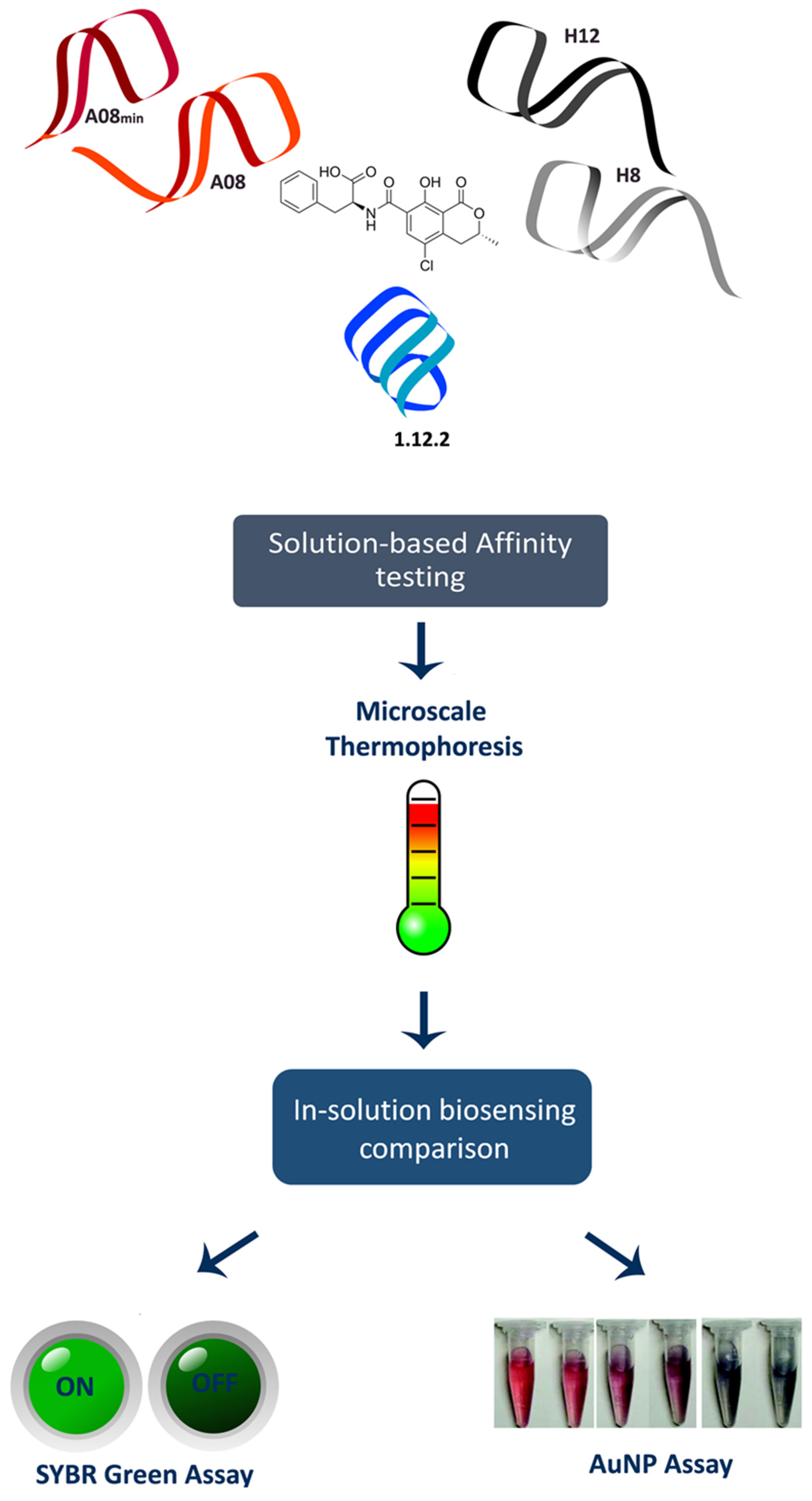
60) M. McKeague, R. Velu, A. De Girolamo, S. Valenzano, M. Pascale, M.K. Smith, M. C. DeRosa* “Comparison of In-Solution Biorecognition Properties of Aptamers against Ochratoxin A” Toxins 2017 8, 336-340.
59) E. M. Hassan, W.G. Willmore, M.C. DeRosa* “Aptamers: Promising Tools for the Detection of Circulating Tumor Cells” Nucl. Acid. Ther. 2016, 26, 335-347.
58) E.M. McConnell, R. Bolzon, M. C. DeRosa* “pHAST (pH-driven Aptamer Switch for Thrombin) catch-and-release of target protein” Bioconj. Chem., 2016, 27, 1493-1499.

57) S. Valenzano*, A. De Girolamo*, M. C. De Rosa*, M. McKeague, R. Schena, M. Pascale “Screening and identification of DNA aptamers to tyramine using in vitro selection and high-throughput sequencing” ACS Comb. Sci. 2016, 18, 302-313. (Cover)

56) A. Ruscito, M .C. DeRosa* “Small-Molecule Binding Aptamers: Selection Strategies, Characterization, and Applications” Front. Chem. 2016, 4, 14. (Invited, Special Issue)
55) A. Ruscito, M. Smith, D. Goudreau, M. C. DeRosa* “Current status and future prospects of aptamer-based mycotoxin detection” J. A. O. A. C. Int. 2016, 99, 865-877.
54) E. Mastronardi, P. K. Tsae, X. Zhang, A. Foster, Y. Sultan, M. C. DeRosa* “Preparation and Characterization of Aptamer-Polyelectrolyte Films and Microcapsules for Biosensing and Delivery Applications.” Methods 2016, 97, 75–87. (Invited, Special Issue).

53) F. Costantini, C. Sberna, G. Petrucci, M. Reverberi, F. Domenici, C. Fanelli, C. Manetti, G. de Cesare, M. C. DeRosa, A. Nascetti, D. Caputo, “Aptamer-based sandwich assay for on chip detection of Ochratoxin A by an array of amorphous silicon photosensors” Sens. Act. B: Chem. 2016, 230, 31-39.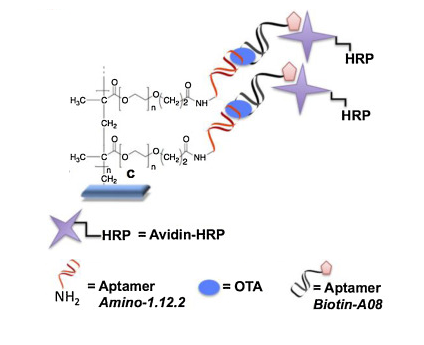
52) M. McKeague, E. M. McConnell, J. Cruz-Toledo, E. Bernard, A. Foster, E. Mastronardi, X. Zhang, M. Beking, T. Francis, A. Giamberardino, A. Cabecinha, A. Ruscito, R. Aranda-Rodriguez, M. Dumontier*, M. C. DeRosa* “Analysis of In Vitro Aptamer Selection Parameters.” J. Mol. Evol. 2015, 81, 150-161. (Invited, Special Issue)

51) C. M. Monreal, M. C. DeRosa, S. C. Mallubhotla, P. S. Bindraban, C. Dimkpa “Nanotechnologies for increasing the crop use efficiency of fertilizer- micronutrients” Biol. Fert. Soils. 2015, 1-15.
50) G. Luka, A. Ahmadi, H. Najjaran, E. Alocilja, M. C. DeRosa, K. Wolthers, A. Malki, H. Aziz, A. Althani, M. Hoorfar “Microfluidics integrated biosensors: A leading technology towards lab-on-a-chip and sensing applications” Sensors 2015, 15, 30011-30031.
49) N. R. Frost, M. McKeague, D. Falcioni, M. C. DeRosa* “An in solution assay for interrogation of affinity and rational minimer design for small molecule-binding aptamers.” Analyst, 2015, 140, 6643-6651.
48) M. McKeague, A. De Girolamo, S. Valenzano, M. Pascale, A. Ruscito, R. Velu, N. R Frost, K. Hill, M. Smith, E. M. McConnell, M. C. DeRosa* “Comprehensive Analytical Comparison of Strategies Used for Small Molecule Aptamer Evaluation.” Anal. Chem. 2015, 87, 8606-8612. (Chosen for ACS Editor’s Choice)

47) R. Velu, N. Frost, M. C. DeRosa* “Linkage Inversion Assembled Nano-Aptasensors (LIANAs) for Turn-On Fluorescence Detection” Chem. Comm. 2015, 51, 14346-14349.
46) C. Mattice, M. C. DeRosa* “Status and Prospects of Aptamers as Drug Components” BioDrugs, 2015, 29, 151-165 (Invited review)
45) P. Tsae, M. C. DeRosa* “Outlook for Aptamers After Twenty Five Years” Curr. Top. Med. Chem. 2015, 15, 1153-1159. (Invited review).

44) R. Walsh, U. Ho, X.-L. Wang, M. C. DeRosa* “Selective Dopamine Detection Using Aptamer-Functionalized Glassy Carbon Electrodes” Can. J. Chem. 2015, 93, 572-577.

43) R. H. Griffin, E. M. McConnell, M. C. DeRosa*, N. G. Tarr* “MOS Testbed for the characterization of Targeted Alpha Therapy Pharmaceuticals.” IEEE Sensors Journal 2015, 15, 1690-1696. (cover)
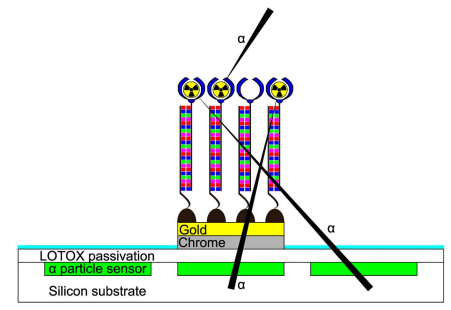
42) E. B. Bernard, K. C. Nguyen, M. C. DeRosa, A. F. Tayabali, R. Aranda-Rodriguez “Development of a Bead-Based Aptamer/Antibody Detection System for C-Reactive Protein. Anal. Biochem. 2015, 472, 67-74.
41) E. M. McConnell, M. R. Holahan, M. C. DeRosa “Opportunities and Limitations for Aptamers in the Central Nervous System” Nucl. Acids Ther. 2014 24, 388-404. (Invited Review).
40) M. McKeague and M.C. DeRosa “Aptamers and SELEX: Tools for the Development of Transformative Molecular Recognition Technology” Aptamers and Synthetic Antibodies 2014 1, 12-16. (Invited review)
39) M. McKeague, V. Ranganathan, K. Hill, V. Bardozy, T. Mezaros, M. C. DeRosa “Selection and characterization of novel DNA aptamers for label-free fluorescence biosensing of ochratoxin A” Toxins 2014 6, 2435-2452.
38) A. Foster, M.C. DeRosa. “Development of a Biocompatible Layer-by-Layer Film System using Aptamer Technology for Smart Material Applications” Polymers 2014 6, 1631-1654.
37) E. Mastronardi, A. Foster, X. Zhang, M. C. DeRosa “Smart materials based on DNA aptamers: Taking aptasensing to the next level.” Sensors 2014 14, 3156-3171. (Invited Review)
36) F. Matus, C. Monreal, M. Lefebvre, S-S. Wu, R. Desjardins, M. C. DeRosa “Producing Isotopically Enriched Plant, Soil Solution and Rhizosphere Soil Materials Over a Few Hours” Comm. Soil Sci Plant Anal. 2014, 45, 865-880.
35) R. H. Griffin, O. Mozenson, M.A. Beking, M. C. DeRosa, G. Lopinski, N.G. Tarr “Quantitative radiolabeled biomolecule detection using a functionalized CMOS sensor” IEEE Trans. Nucl. Sci., 2014, 61, 1112-1117.
34) S. Smiley, M. C. DeRosa, B. Blais “Immobilization of DNA Aptamers on Polyester Cloth for Antigen Detection by Dot Blot Immunoenzymatic Assay (Aptablot)” J. Nucleic Acids 2013, 2013¸963542.
33) X. Zhang, D. Chabot, Y. Sultan, C. Monreal, M. C. De Rosa “Target-molecule-triggered rupture of aptamer-encapsulated polyelectrolyte microcapsules.” ACS Appl. Mater. Interfaces 2013, 5, 5500-5507.
32) A. Giamberardino, M. Labib, E. Hassan, J. A. Tetro, S. Springthorpe, S. A. Sattar, M. V. Berezovski and M. C. DeRosa “Ultrasensitive Norovirus Detection using DNA Aptasensor Technology.” PLoS One, 2013, 8, e79087.
31) M. McKeague, A. Foster, Y. Miguel, A. Giamberardino, C. Verdin, J.Y.S. Chan, M.C. DeRosa “Development of nucleic acid probes for direct and selective homocysteine detection in human serum” RSC Advances 2013, 3i, 24415-24422.
30) J. Albert, S. Lepinay, C. Caucheteur, M. C. DeRosa “High resolution grating-assisted surface Plasmon resonance fiber optic biosensor” Methods 2013, 63, 239-254.
29) M. McKeague and M. C. DeRosa, “Challenges and Opportunities for Small Molecule Aptamer Development,” J. Nucleic Acids, 2012, 2012,748913. (invited review).
28) Bernard, E. D., Beking, M. A., Rajamanickam, K., Tsai, E. C. and M. C. DeRosa “Target binding improves relaxivity in aptamer-gadolinium conjugates.” J. Biol. Inorg. Chem 2012, 17, 1159-1175.
27) J. Cruz-Toledo,M. McKeague, X. Zhang, A. Giamberardino, E. McConnell, T. Francis, M. C. DeRosa, and M. Dumontier “Aptamer base: a collaborative knowledge base to describe aptamers and SELEX experiments” Database 2012, bas006.
26) B. Xi, I. P. Liu, G. L. Xu, M. M. Choudhuri, M. C. DeRosa, R. J. Crutchley, T. Ren “Modulation of Electronic Couplings within Ru2–Polyyne Frameworks” J. Am. Chem. Soc. 2011, 133, 15094–15104.
25) Y. Shevchenko, T. J. Francis, D. A. Blair, R. Walsh, M. C. DeRosa, J. Albert “In situ biosensing with a surface plasmon resonance fiber grating aptasensor” Anal. Chem. 2011, 83, 7027-7034.
24) M. R. Holahan*, D. Madularu, E. M. McConnell, R. Walsh, M. C. DeRosa* “Intra-accumbens injection of a dopamine aptamer abates MK-801-induced cognitive dysfunction in a model of schizophrenia” PLoS ONE 2011, 6, e22239.
23) Y. Sultan and M. C. DeRosa* “Target Binding influences permeability in aptamer-polyelectrolyte microcapsules” Small 2011, 7, 1219-1226.
22) A. De Girolamo*, M. McKeague, J.D. Miller, M.C. DeRosa, A. Visconti* “Determination of ochratoxin A in wheat after clean-up through a DNA aptamer-based solid phase extraction column” Food Chem. 2011, 127, 1378–1384.
21) M. McKeague, C. R. Bradley, A. DeGirolamo, A. Visconti, J. David Miller, M. C. DeRosa* “Screening and Initial Binding Assessment of Fumonisin B1 Aptamers” Int. J. Mol. Sci. 2010, 11, 4864-4881. (invited)
20) X.-M. Luo, M. McKeague, S. Pitre, M. Dumontier, J. Green, A. Golshani, M. C. DeRosa*, F. Dehne* “Computational Approaches Towards the Design of Pools for the In Vitro Selection Of Complex Aptamers” RNA 2010, 16, 2252-2262. Highlighted by the Faculty of 1000
19) M. C. DeRosa *, C. Monreal, M. Schnitzer, R. Walsh, Y. Sultan “Prospects for Nanotechnology in Fertilizers” Nature Nano. 2010, 5, 91. Highlighted by the Chemical and Engineering News
18) J. A. Lee, M.C. DeRosa* “A pH-driven DNA switch based on the A+•G mispair” Chem. Commun. 2010, 46, 418 – 420.
17) R. Walsh, M. C. DeRosa* “Retention of Function in the DNA homolog of the RNA aptamer” Biochem. Biophys. Res. Commun. 2009, 388, 732-735.
16) Y. Sultan, R. Walsh, C. Monreal, M.C. DeRosa* “Preparation of Functional Aptamer Films by Layer-by-Layer Self-Assembly” Biomacromolecules 2009, 10, 1149-1154.